Spanish Version
Spanish Version
|
|
|
|
MEMBRANE PROTEINS
There are two types: (1) Integral proteins: are those that cross the membrane and appear on both sides of the phospholipid bilayer. Most of these proteins are glycoproteins, proteins that have several monosaccharides attached. The carbohydrate part of the molecule is always facing to the cell exterior, and (2) Peripheral proteins: these are not extended across the width of the bilayer but are attached to its inner or outer surfaces and are easily separated from it. The nature of Membrane Proteins determines its function: Channels: whole proteins (usually glycoproteins) which act as pores by which certain substances can enter or leave the cell, Carriers: are proteins that change shape to give way to certain products, Receptors: are integral proteins that recognize certain molecules to which they are attached or fixed. These proteins can identify a hormone, a neurotransmitter or a nutrient that is important for the cell function. The molecule that binds to the receptor is called ligand, Enzymes: can be integral or peripheral and serve to catalyze reactions at the surface of the membrane, Cytoskeleton anchors: are peripheral proteins found in the part of the membrane cytosol which serve to fix the cytoskeleton filaments, Cell identity markers: are glycoproteins and glycolipids characteristic of each individual and which allow the identification of cells from another organism. For example, blood cells have ABO markers that make possible that in blood transfusion only blood of the same type can be compatible. Being in the exterior, the carbohydrate chains of glycoproteins and glycolipids form a kind of cover called glicocalix.  +info
+infoANTIMICROBIAL PEPTIDES On february 2017 the World Health Organization issued an alert for the indiscriminate use of conventional antibiotics which has facilitated the resistance of various bacteria to multiple drugs. As the pharmaceutical industry is generating less and less antibiotics due to the lack of investment in research, and at the same time, more and more strains of pathogens are emerging, such as the superbugs resistant to the drugs currently available, the therapeutic options are being exhausted very quickly. Membrane permeabilizing antimicrobial peptides (AMPs) are good candidates to deal with this crisis since they act without high specificity towards a given protein target, which reduces the likelihood of induced resistance. Understanding the AMPs permeabilization mechanism on the bacterial membrane is crucial for the development of new AMPs as useful antimicrobial agents. In order to understand the molecular mechanism and specificity of AMPs for their cellular target, my research group is performing molecular dynamics simulations (MD) of molecular models of complexes formed by AMPs molecules, isolated from scorpion venoms, in the presence of patches of membranes of different lipid composition in aqueous solution and in physiological conditions.  +info
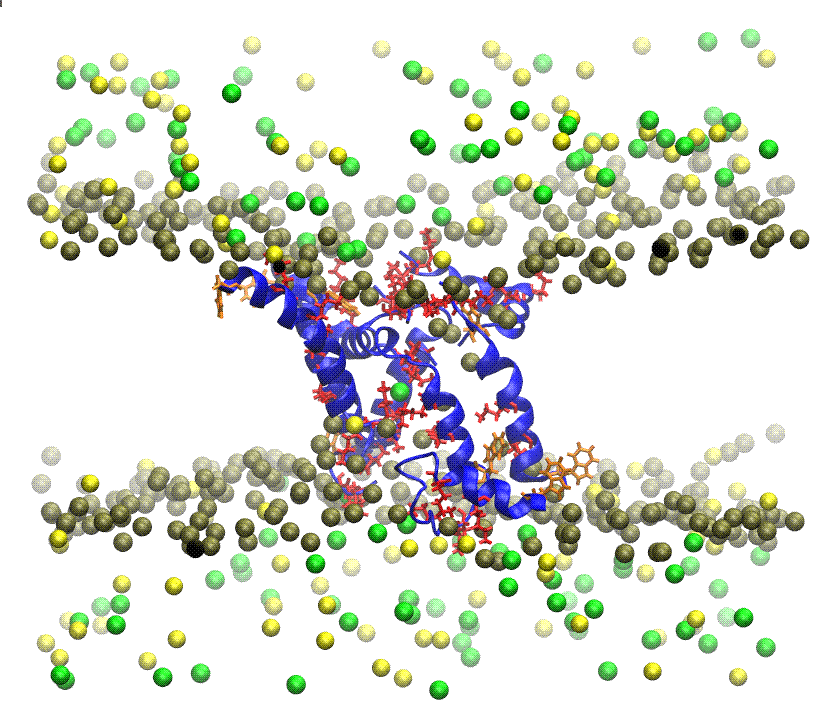
+infoIONIC CHANNELS The term ion channels is applied to certain macromolecular elements, of protein nature, inserted into the cell membranes. In excitable cells their function is to produce and transduce electrical signals. In other cell types these transmembrane proteins form pores and control the passage of specific ions through the membranes. These channels play an important role in the regulation of transepithelial transport of water and salts, and cellular volume and pH, and act as cell signaling pathways. In general, ion channels act as open and closed gates depending on external stimuli, although some toxic substances may deactivate its natural function. In my research group we are interested in studying through MD the ion channel conformational changes that occur when neurotoxins of animal origin, such as those found in scorpion venoms, exert their action on voltage-dependent sodium and potassium channels.  +info
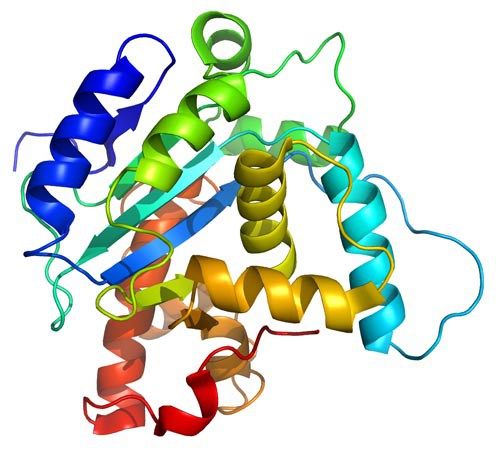
+infoPROTEIN FOLDING When the tertiary structure of a protein has not been determined experimentally, one can try to construct a three-dimensional model from its amino acid sequence. The principle of Anfinsen states that all the information required to specify the structure of a protein is encoded in its amino acid sequence. The objective of the tertiary structure of proteins prediction is to estimate the spatial position of each and every one of the atoms of the protein molecule using computational methods and the information of the protein´s primary structure (i.e. the amino acid sequence). This is one of the most difficult challenges facing Bioinformatics and some research groups have defined it as "the holy grail of bioinformatics", also known as the "Protein Folding Problem". My research group uses ab initio, or de novo, methods to construct a 3D model of a protein. In these methods, only the amino acid sequence of the protein is known, and based exclusively on physical principles (an energy function) the determination of the lowest energy conformation, that represents the global minimum of this function, is obtained using methods of numerical optimization used for NP-hard problems such as: Heuristic Algorithms. In the past we have used heuristics such as Tabu Search and Simulated Annealing. Bioinspired Algorithms. Bio-inspired algorithms are methods of optimization based on some evolutive metaphor or by simulation of social behaviors of insects or humans. Its use has become popular to perform efficient searches and as optimization techniques because they are very effective in solving very difficult optimization problems. Among them we have the Genetic Algorithms, the Ant Colony and Particle Swarm optimization. Knowledged Based Potentials. While Potentials of Mean Force can in principle be calculated from a field of physical force, the basic philosophy of the Knowledge Based Approach is to approximate these quantities by the statistical analysis of known protein structures, sometimes referred to as the "Boltzmann inverse law". Typically, a database that contains a set of known native structures from high-resolution data is used, which can include a number of decoys depending on the application. The database that is used for the construction of knowledge-based potentials will typically include variations both structural and sequential.  +info
+infoENANTIODIFFERENCIATION We are also interested in knowing the molecular basis of the enantiodifferentiation process in chiral drugs. Our interest in the matter emerged when we learnt that between 1959 and 1964 the drug called Thalidomide (a chiral molecule) was prescribed to treat anxiety, insomnia and nausea in pregnant women. Use of this drug caused terrible birth defects. Only in Europe were born more than 10,000 seriously deformed children, many of them without legs or arms, because their mothers had taken the drug at the beginning of their pregnancy. It was later discovered that its R isomer has the teragenic effect, while the S isomer has the desired sedative effect. Today many drugs available on the market are chiral. To understand the molecular basis of the enantiodifferentiation process our research group applies the technique MD on cyclodextrin inclusion complexes as model systems of chiral receptors.  +info
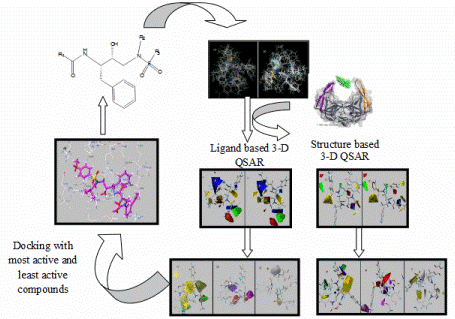
+infoQSAR Models of Quantitative Structure-Activity Relationship (QSAR models) are regression models or classification used in chemical and biological sciences and engineering. Like other regression models, the QSAR regression models relate a set of predictor variables (X) with the power of the response variable (Y), while the QSAR classification models relate the predictor variables to a categorical value of the response variable. In QSAR modeling, the so called predictors consist of physical-chemical properties or theoretical molecular descriptors of the chemical substances. The QSAR response variable could be the biological activity of the chemical moities. The QSAR models first summarize an alleged relationship between chemical structure and biological activity in a set of substances chemical data. And secondly, the QSAR models predict the activities of new chemicals. My research group has been involved in the formulation of QSAR models for compounds with different biological properties such as antinociceptive, antibacterial, human serum albumin inhibitors, vasopressin inhibitors and carcinogens among others.  +info
+infoFREE ENERGY The formation of complexes in which biological molecules participate is not an exclusive phenomenon of Pharmacology, but is contemplated in other branches of Biology, such as Biochemistry (e.g. Michaelis complexes between enzymes and substrates) or Immunology (e.g. antigen-antibody complexes), and and those proper of Chemistry, where interactions between "guest" and "host" molecules have opened new perspectives in the denominated Supramolecular Chemistry. Any pharmacological action has its beginning in the formation of a complex between the drug molecule and its receptor site in a biological macromolecule, mostly proteins. Therefore, the specificity of the response to a given drug is determined largely by the ability of recognizing it as agonist or antagonist and being able to evoke a response or not. For the study of ligand-receptor interactions, we used the molecular dynamics methods for the calculation of the free energy of association (ΔG) such as: (a) Free Energy Perturbation, (d) Umbrella Sampling, (c) Thermodynamic Perturbation, (d) Biased Sampling and its variations. For these and other techniques you can consult the Free Energy Computations book.  +info
+info |
|
Copyright © Ramón Garduño-Juárez. All Rights Reserved. |
 E-mail: | Mayo 2017
E-mail: | Mayo 2017